Experimenting with Deep Neural Networks for X-Ray Image Segmentation
Deep neural networks possess a variety of possibilities for improving medical image segmentation. This article shares some of the results of a research conducted by our partners at the Biomedical Image Analysis Department of the United Institute of Informatics Problems, National Academy of Sciences of Belarus. The study aimed at examining the potential of deep learning and encoder-decoder convolutional neural networks for lung image segmentation.
Source data
The training data set consisted of 354 chest X-ray images accompanied by the lung masks obtained through manual segmentation. Two different image sources were used:
- 107 images from the Belarus tuberculosis portal manually segmented during the preliminary phase of this project
- 247 images from the JSRT database
Examples of the original images and corresponding lung masks are illustrated in the following figure.
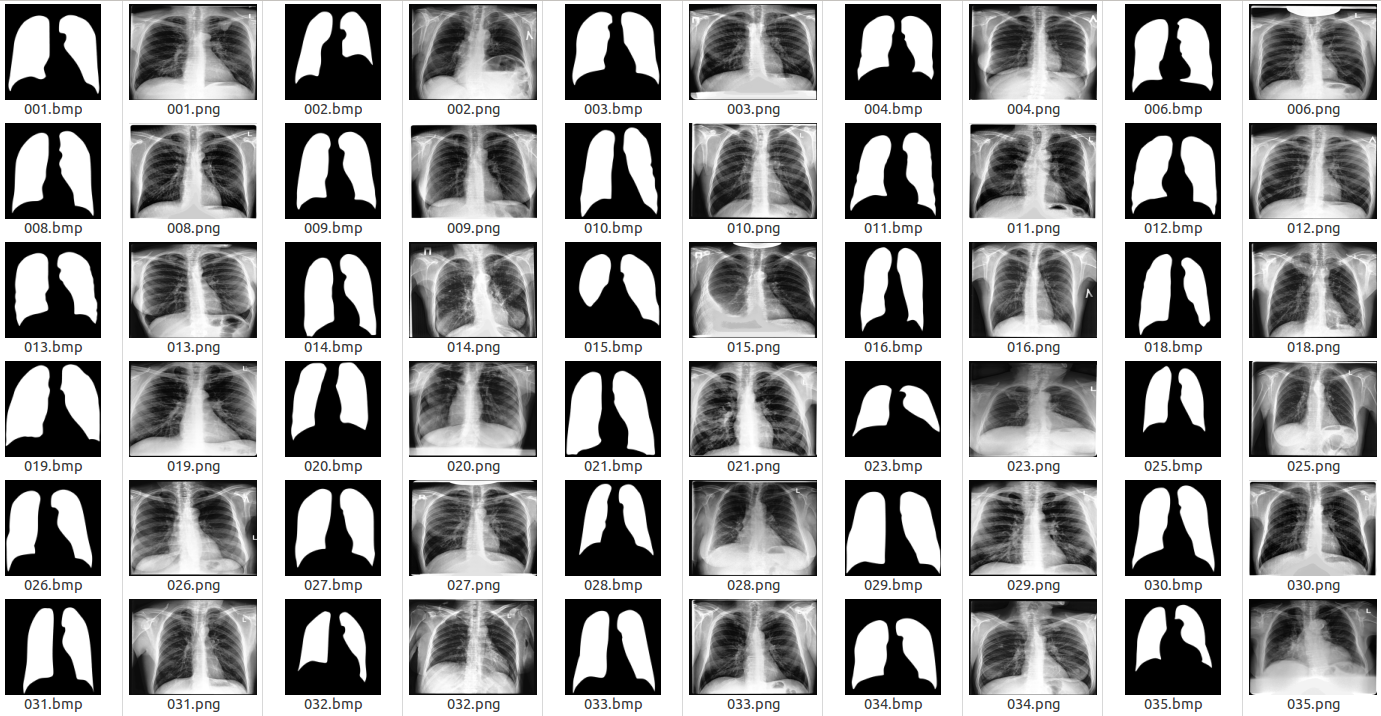 Examples of X-ray images and corresponding lung masks
Examples of X-ray images and corresponding lung masks
Network architecture and parameters
In the figure below, you can find the neural network architecture that was used during the study.
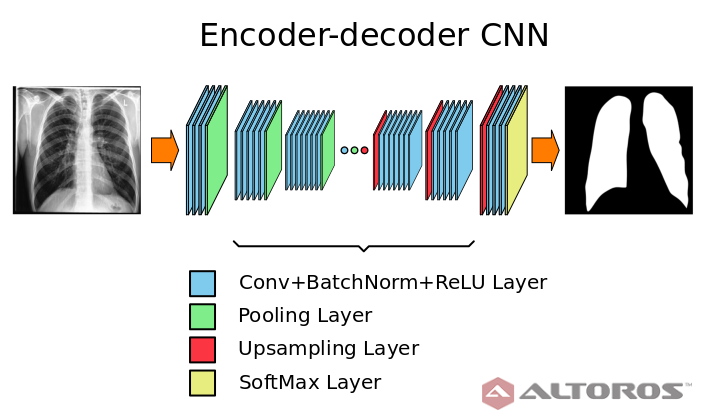 A simplified scheme of the encoder-decoder neural network architecture
A simplified scheme of the encoder-decoder neural network architectureThe network had a typical deep architecture with the following key elements:
- 26 convolutional layers
- 25 batch normalization layers
- 25 ReLU layers
- 5 upsampling layers
All experiments and testing were performed using the Caffe framework. The input and output network fragments are illustrated in the figure below.
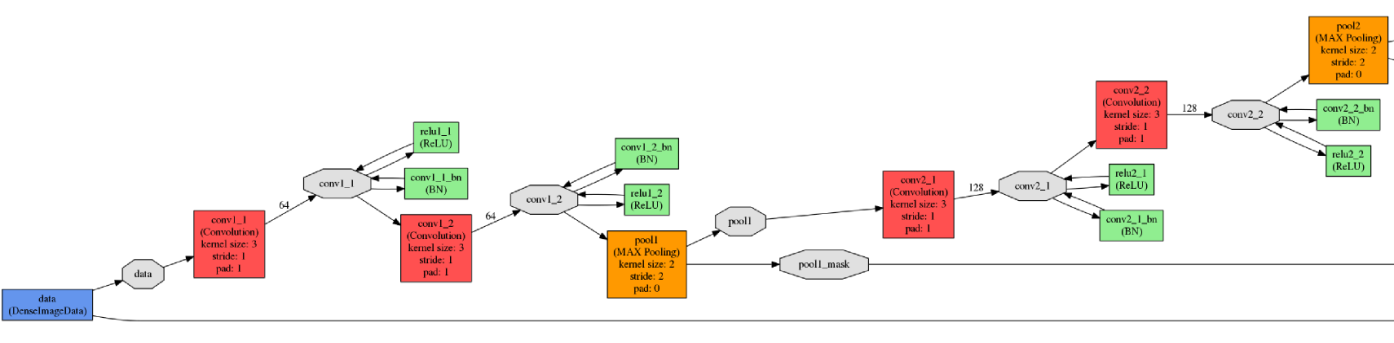
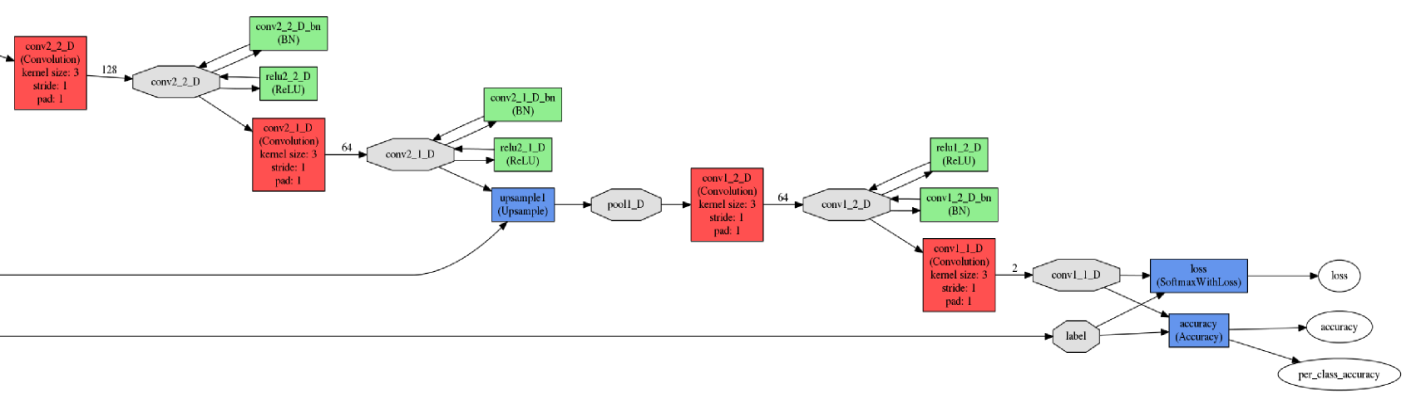 The input (top) and output (bottom) network elements
The input (top) and output (bottom) network elementsThe neural network was trained on the NVIDIA TITAN X graphics processor with 12 GB of GDDR5 memory. The network training parameters were as follows.
- Batch size: 6
- Caffe solver: SGD
- Number of iterations: 5,000
- Number of epochs: 85
The total time of the neural network training was approximately three hours. During the training stage, the neural network used approximately 11 GB of GPU memory.
Brief analysis
As a result, the segmentation accuracy was assessed by comparing the automatically obtained lung areas with the manual version using Dice’s coefficient, which is calculated as shown in the formula below.
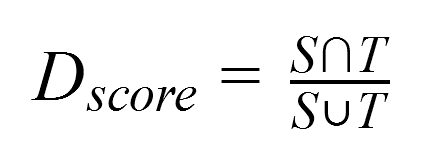
where:
Tis the lung area resulted from manual segmentation and considered as ground truth.Sis the area obtained through automatic segmentation using the neural network.
During the testing stage, the average accuracy was estimated as 0.962 (the minimum score value was 0.926 and the maximum score value was 0.974) with the standard deviation of 0.008.
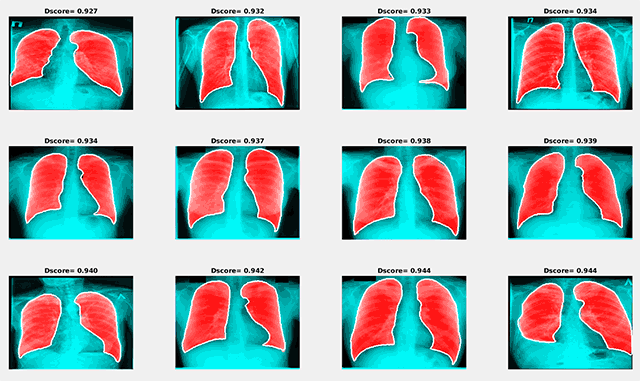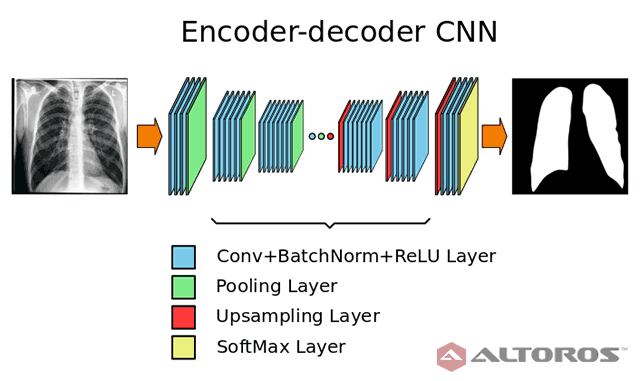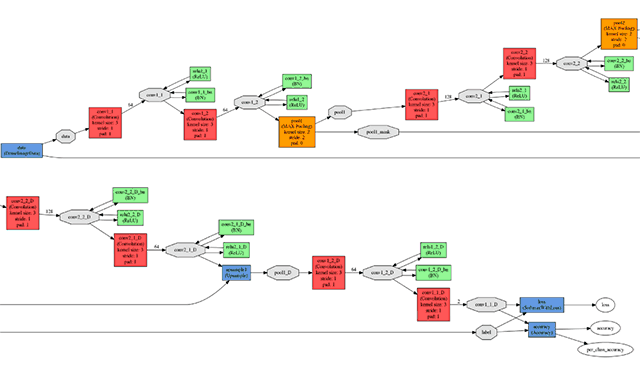
Examples of the best and worst segmentation results are given in the following figures. The red area in the image below presents the results of segmentation using the trained neural network, and the white line shows the ground truth lung mask boundary.
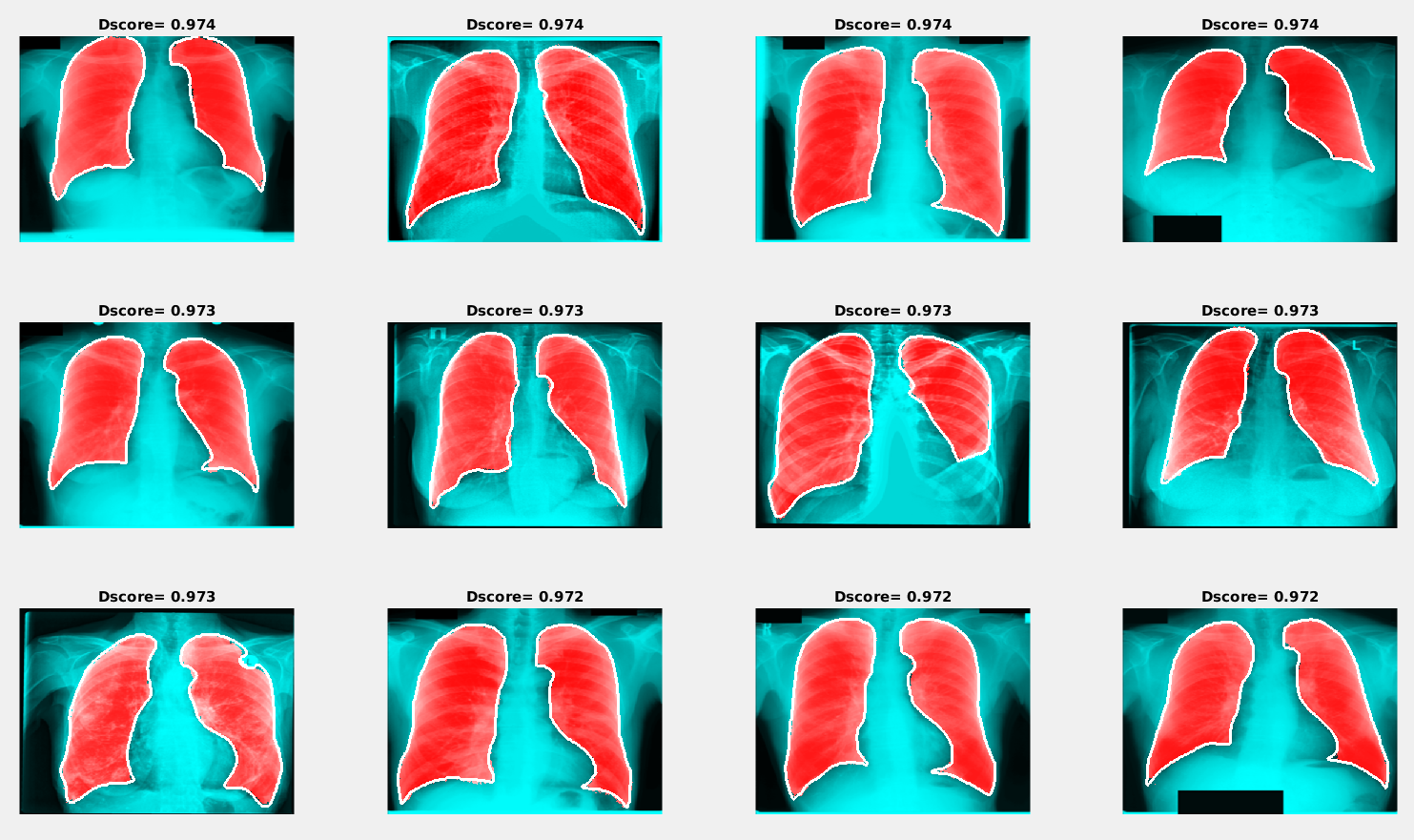 Examples of segmentation results with the maximum Dice score
Examples of segmentation results with the maximum Dice scoreSimilar to the previous image, the red area in the figure below shows the results of segmentation using the trained neural network, and the white line presents the ground truth lung mask boundary.
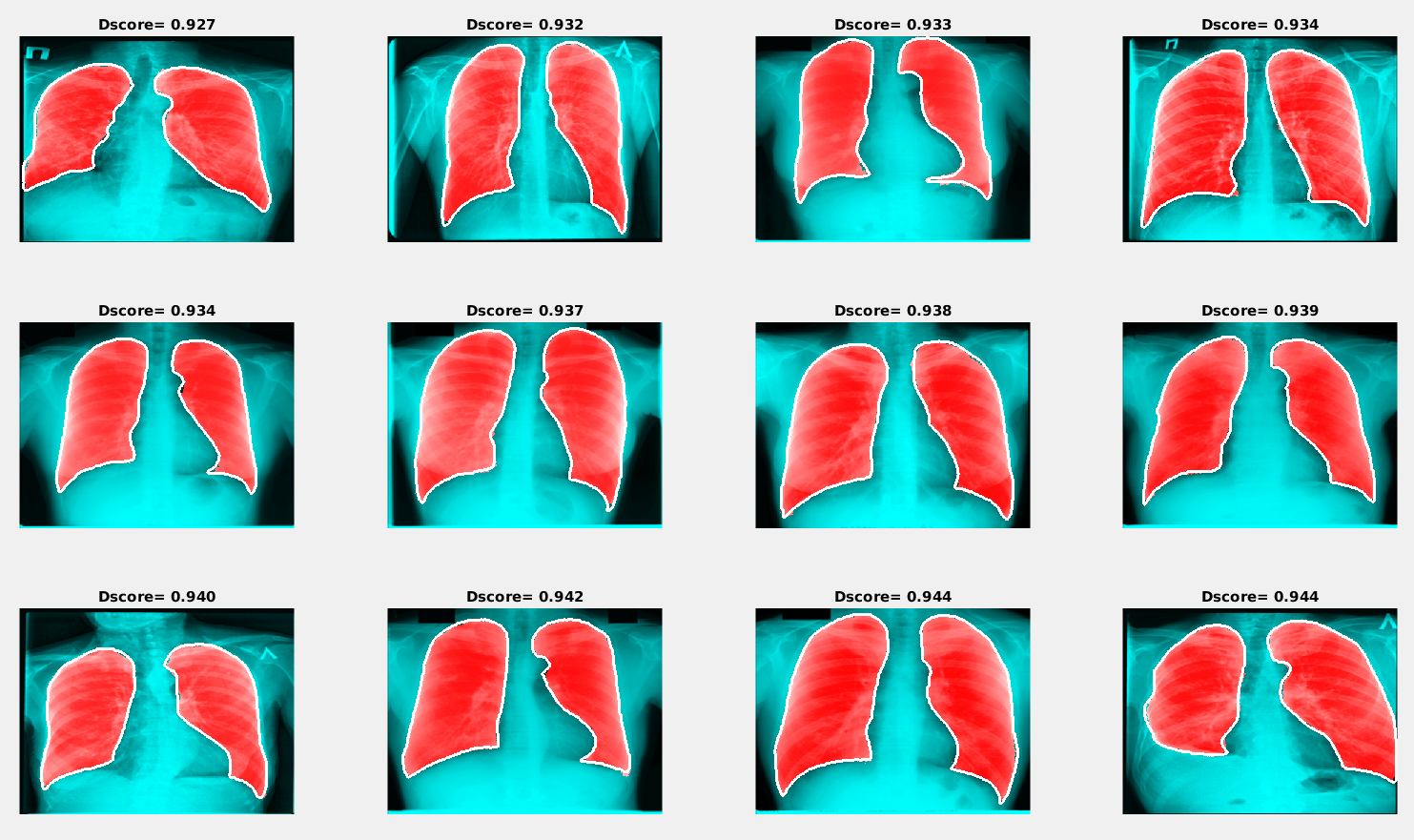 Examples of segmentation results with the minimum Dice score
Examples of segmentation results with the minimum Dice scoreThe results obtained during this study have demonstrated that encoder-decoder convolutional neural networks can be considered as a promising tool for automatic lung segmentation in large-scale projects. For more details about the conducted research, read the paper, “Lung Image Segmentation Using Deep Learning Methods and Convolutional Neural Networks” (PDF).
The described scenario was implemented with the Caffe deep learning framework. If you have tried to use Deeplearning4j, TensorFlow, Theano, or Torch for similar purposes, share your experience in the comments.
Further reading
- Recurrent Neural Networks: Classifying Diagnoses with Long Short-Term Memory
- TensorFlow and OpenPOWER Driving Faster Cancer Recognition and Diagnosis
- Using Convolutional Neural Networks and TensorFlow for Image Classification